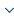1. Introduction
In recent years, the incidence and mortality rate of lung cancer is the firstin China, and lung nodules are the early stage of lung cancer. With the continuous development of the era of big data and the rapid growth of image data, image data of about 2PB or more in medical images generated by national hospitals each year is used. Therefore, this paper’s research significance is about how to utilize a large amount of image data to share diagnostic reports and help doctors’ auxiliary diagnosis. Content-based medical image retrieval (CBMIR) is a technique that retrieves similar images from the database by extracting the semantic features of the images. Hence, the correct and comprehensive expression of image semantic information has become the key to image retrieval. There are two main aspects to improve the retrieval performance of CBMIR: first: Extracting medical image features.Feature extraction of medical images. Sufficient characterization can improve the accuracy of the retrieval system. In the traditional content-based image retrieval system, because we extract features manually, these features are subjective and incomplete, and resulting in low retrieval accuracy. In this paper, we use the fully convolutional dense network (FCDN) to automatically extract objective and comprehensive features from medical images. Second:Retrieving the performance of medical images.Medical image retrievalefficiency.Thedescription of image information will generate a high dimensional feature vector. It can be directly used for similarity search, but reduces the performance of image retrieval. In [1], a learning framework based on DCNN is designed for feature extraction of medical images. Although these frameworks based on deep learning improve the representation of image features, there are some problems that affect its scalability and practicality, such as the complex design of deep neural networks and a long time for training.
Designing an advanced neural network structure is one of the most challenging and effective ways to improve image feature extraction. AlexNet [2] and VGG [3] are the two most important network structures for deep convolutional neural network. They demonstrated that building a deeper network with tiny convolution kernels is an effective way to improve neural network learningability. After that, they proposed a ResNet structure [4-5], which reduced the optimization difficulty and further pushed the depth of neural network to hundreds of layers. Since then, different types of ResNet have emerged, devoted to establishing a more efficient micro-block internal structure or exploring how to use residual connections [6]. Recently, in [7] proposed a dense network (DenseNet) in which input features and output features are connected in parallel using dense blocks, which is also the major difference from the residual network. DenseNet has several advantages: It mitigates the problem of gradient disappearance, enhances feature propagation, and encourages feature reuse. However, the width of densely connected paths increases linearly with increasing depth, if not optimized in the implementation, causes a second increase in the amount of parameters and consumes large amounts of GPU memory compared to the residual network, thus limiting deeper DenseNet to further improve the accuracy. In addition to designing a new network structure, the researchers also tried to re-explore the state-of-the-art architecture. The authors proposed in [8] that the residual path is an important factor to reduce the network optimization difficulty. For DenseNet, in addition to introducing better feature reuse and mitigating the issue of gradient dissipation during training, it also could gain deeper insight into dense networks.
The content-based medical image retrieval mainly includes two parts: feature extraction and hash function. Feature extraction is the cornerstone of retrieval accuracy. Based on the above problems and the characteristics of lung nodules images, this paper proposes a fully convolutional dense network (FCDN). FCDN solves the problem of insufficient feature extraction and image retrieval low accuracy. FCDN is mainly divided into two phases Encoding and Decoding. Therole of Encoding is to extract the rich semantic features of lung nodules images, feature extraction to express image information. The role of Decoding is to restore the size of the input image by up-sampling the feature map with rich semantic information after pooling. In addition, the Joint distance proposed is to add the output of different layers of the network as the final retrieval image features. Different layers extract the information of the images differently.The Joint distance not only expresses the information of lung nodules images, but also improves the retrieval precision. Through the analysis of the experimental results, compared with different hashing methods such as LSH, SH, KSL and so on, the proposed fully convolutional dense network and Joint distance have achieved better results in retrieval performance evaluation.
2. Related Work
2.1. Data Processing
Large-scale annotation datasets with representative data features are crucial for learning models that are more accurate and extendable [9], such as datasets. ImageNet [10] is a very comprehensive database of more than 1, 000 species of over 1.2 million categories natural images. However, there are no large annotated lung nodule image datasets comparable to ImageNet, because the acquisition of medical image data is very difficult and there are fewer annotated tags. This article uses the standard dataset published by the kaggle algorithm design contest Luna2016, including 1186 nodules in 888 cases. Because the source data obtained contains multiple slices in each case, and it is hard to find the specific location of the lung nodules in the slice, it needs image pre-processing. First, mhd format sources data through binary, morphological filtering expansion, closing operation and other operations to generate only the lungs of the image. Figure 1_a(f)shows network input image. Set the CT segmentation threshold to -1200 to 600HU while reducing the CT scan pixel differences for different devices by scaling and interpolation methods. In order to prevent the negative sample from affecting too much of the final result of the experiment, we apply the data-enhanced method to expand the positive sample to a ratio of about 1:1, which sets the positive sample to7116 and the negative sample to 6888, to ensure a balanced sample. Figure 1_b shows different shapes and sizes of lung nodules.
Figure 1
Figure 1.
a. (a)Origin;(b) Binary; (c) Clear-border; (d) Closing; (e) Dilation; (f) Input;b. Lung nodule samples
2.2. The Principle of the Medical Image Retrieval
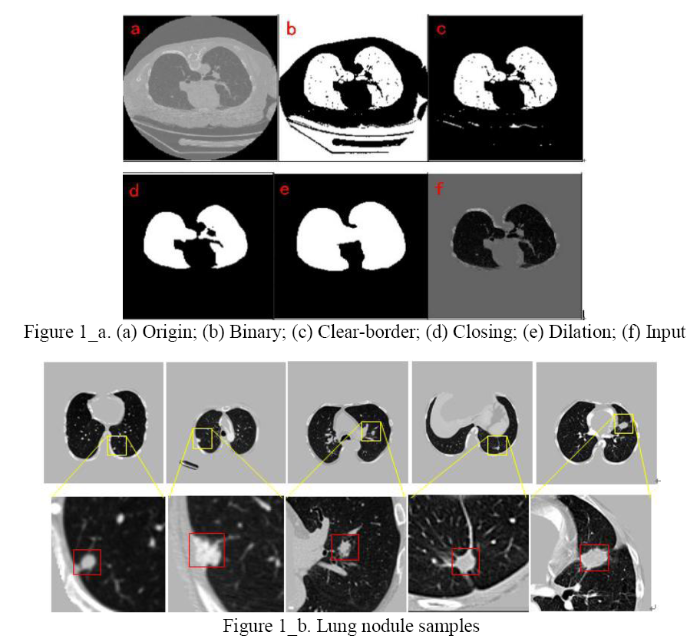
Content-based medical imageretrieval (CBMIR) principle is shown in Figure 2. First, there is a lung nodule image dataset in the database, extracting the characteristics of lung nodules by network structure training and generating image feature vectors.High-dimensional feature vectors have low retrieval efficiency and high memory consumption for images. A hash method is required to map the high-dimensional features of the image into compact binary hash codes. When the image to be retrieved is entered, it is similarly passed through the network to generate a compact binary code. We use the Hamming distance as a measure of similarity. The smaller the hamming distance is, the higher the similarity of the two images is.According to demand, we return retrieval results.
Figure 2.
Figure 2.
Medical image retrieval principle
2.3. Feature Extraction
The key of feature extraction is to obtain the difference image information. If the extracted features cannot distinguish between different medical images, then the feature extraction step will be meaningless. The histogram of gradient (Hog) [11] is a feature descriptor used for object detection in computer vision and image processing. It computes the features by calculating and counting histograms of gradient directions in the local area of the image. In [12], the scale-invariant feature transform (SIFT) algorithm is based on the points of interest of some local appearances on the image and has nothing to do with the size and rotation of the image and eliminates the key points of the unmatched relationship caused by image occlusion and background chaos. These methods have certain limitations on the image information expression. With the development of deep learning, a convolutional neural network (CNN) for multi-layer perception (MLP) to acquire image features is proposed. CNN has some advantages that traditional technologies do not have: good fault tolerance, parallel processing and self-learning capabilities, and high resolution. At the same time, its non-fully connected and weighted sharing reduces the complexity of the network model.
2.4. Hash Method
A great deal of methods has been proposed around hash learning. Although the retrieval efficiency is improved, the loss of binary quantization also results in the lack of the feature information of the image and the decrease of the accuracy [13]. Hash is designed to produce highly compact binary code that is well suited for large-scale image retrieval tasks. Most existing hashing methods transform the image into feature vectors and then perform a separate binarization step to generate the hash code. This two-stage process may produce suboptimal encoding. In [14], a depth residual hash (DRH) is proposed to learn two phases simultaneously and to reduce quantization errors and improve the quality of the hash coding by using hash-related loss and regularization. Recently, a new hash index method was proposed in [15], which is called deep hash fusion index (DHFI). Its purpose is to generate more compact hash codes with more aexpressive ability and distinguishing ability.In the literature, a deep hash subnet of two different architectures is trained and the hash codes generated by the two subnets are merged together to express the image information.
3. Method
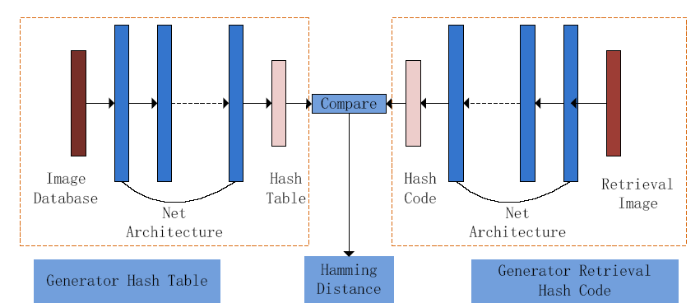
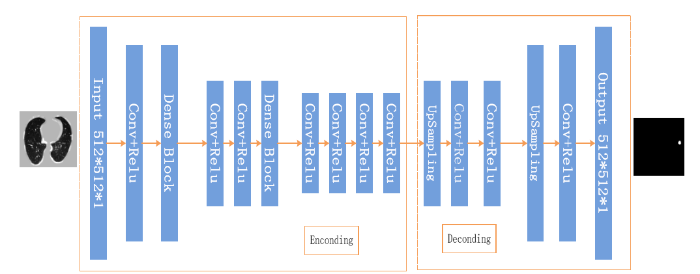
In view of the problems above, combined with the characteristics of datasets and proposed fully convolution dense network (FCDN) shown in Figure 3, the network structure is divided into two parts: Encoding and Decoding. In order to improve the representation of features to ensure maximum information flow between network layers, Encoding join the Dense Block connects all layers with matching feature sizes. The convolutional layer obtains specific image feature input from the previous network layer and passes the feature map to the subsequent network layer.
Figure 3.
Figure 3.
The structure of network
3.1. Encoding
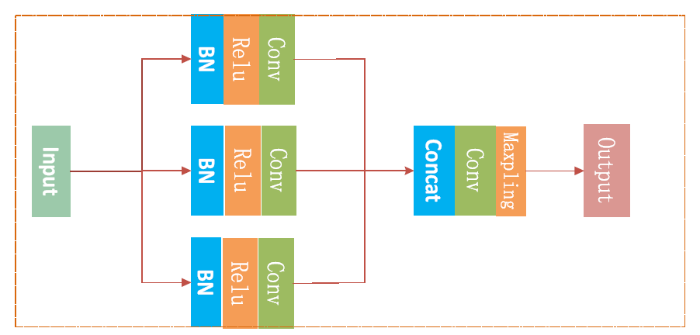
Encoding is the main part of learning the features of lung nodules in the entire network architecture. The network structure of Encoding shown in Figure 3 includes 9 convolutional layers and 3 dense blocks, and each dense block includes 3 layers of BN_Relu_Conv. FCDN to extract richer image information is focused on Dense Block and its basic module content is shown in Figure 4.The module’s main feature is to tap the inherent implicit features of more images to enhance the expression of image. At the same time, to some extent, it reduces the gradient of the training process dissipated. As shown in Figure 4, compared with the residual network, the dense block has better parameter efficiency. In the study of [16], it shows that each layer of ResNet has its own weight and the parameters are too large, which will affect the learning of the model. The advantage of a dense block is that it improves the flow of information throughout the network, making it easy to train. Gradients can be derived directly from the loss function and the original input signal at each layer, which helps training deeper network architectures.
Figure 4.
Figure 4.
The dense block
3.2. Decoding
In the Encoding process, the function of convolution is to extract features [17], and the same convolution will not change the image size. The Decoding process also uses the same convolution, but the role of convolution is upSampling image rich information so that MaxPooling lost information can be learned by Decoding, and can ultimately restore theinputto the same size as the image. The common layer of the network framework has 3 layers of MaxPooling, and 1 layer of MaxPooling for each dense block layer. In the Decoding section there are 6 upSampling to restore the image to the original size. In order to prevent the upSampling process from changing the characteristic information of the image, we use thelinear interpolation method [18]. Finally, the output is showninFigure 3.
3.3. Batch Normalization
${{\mu }_{B}}$ Means the mean value of each batch and the denominator refers to the standard deviation of each batch. Direct use of (1) may undermine the network structure of the characteristic distribution, so the introduction of parameters γ andβare used to maintain the model’s ability to express. It is shown in Equation (2).
The algorithm is applied to the network structure of this paper. After the input image passes the encoding layer, the image information is transformed into multiple feature maps. Therefore, a pair of learning parameters for each feature map is obtained through the parameter sharing strategy: γ,β.The Feature map will do the normalization. In addition, the activation function of the network structure uses the ReLu function to nonlinearly map the output of the convolutional layer. Compared with the sigmoid function, ReLu converges fast and overcomes the gradient disappearance.
3.4. Loss Function
The paper defines two loss functions. Through the characteristics of lung nodule image datasets, the output of FCDN network is considered a two classification problem, so the output loss of FCDN network model is binary cross entropy. In addition, the lung nodule image retrieval function in the pixel area, the prediction of each pixel on the image, and the actual value of the loss of error and sum are defined as Pixel-Area MSE Loss. The total loss function is shown in Equation (3).
In Equation (3),${{L}_{total}}$ represents the overall loss function, binary cross entropy is referred to as${{L}_{bc}}$, Pixel-Area MSE Loss is referred to as ${{L}_{pa}}$,$\lambda$ is the coefficient of ${{L}_{pa}}$, the purpose is to adapt to the total loss function.
3.4.1. Binary Cross Entropy Loss
When training in the network, the quadratic cost function is used as the loss function. The larger the error between the predicted value and the actual value, the smaller the adjustment of the parameter is and the slower the training is. Overcome this problem by increasing the convergence speed of network training. In this paper, the FCDN network model uses cross-entropy as a loss function. Cross entropy is used to evaluate the difference between the probability distribution and the true distribution obtained in the network training process, We reduce the cross-entropy loss to improve the prediction accuracy of the network model.To reduce over-fitting, and avoid local optima, the loss function joins the L2 regularization. The Equation (4) and Equation (5) is as follows.
nrepresents the number of samples in the training dataset and $\eta $ is a parameter of the L2 regularization. The role is to regulate the importance of L2. In Equation (6),${{y}_{j}}$ represents the probability that y equals 1 under ${{x}_{j}}$ condition.
where ${{L}_{0}}=-\frac{1}{n}\sum\limits_{{{x}_{j}}}{[{{y}_{j}}In(h({{x}_{j}}))+(1-{{y}_{j}})In(1-h({{x}_{j}}))]}$,Equation (7) is equivalent to the equation listed below.
The gradient of weight and bias is shown in Equation (8) and Equation (9).
In Equation (10) and Equation (11), m refers to the stochastic gradient descent batch size, and $\eta $refers to the parameter update is the step size.
3.4.2. Pixel-Wise MSE Loss
Mean square error (MSE) is commonly used as a loss function to assess the performance of the model. It refers to the expected value of the square of the difference between the estimated value and the true value. The smaller the value of MSE, the more accurate the model describes the experimental data. The sum of the mean square error of pixel domain was proposed. The MSE is calculated for each pixel through an end-to-end training network structure. When the total loss of the pixel domain is the smallest, the retrieval accuracy of lung nodules is the highest.It is shown in Equation (12).
W represents a wide lung nodule image, H represents a high lung nodule image, ${{P}^{HR}}$ represents a predicted value of current pixel, ${{P}^{TL}}$ represents a label value of current pixel, and LMESrepresents sum of MES an image. The smaller theLMES, the higher the similarity of the images.
3.5. Hash Function
There are three major steps in hash function learning. The first step is to learn image features and hash functions through the training model of the network. The second step is to generate the optimal target hash code from the tag information. The third step is to map the image pixels to a compact binary code using a hash function and perform an image search. If the length of the generated hash code is k, the hash function is shown Equation (13).
x is the feature of the lung nodule image. Let b equal to 0.The Equation(14) is simplified as
In Equation (15), $\min D$ Denotes the minimum Hamming distance, ${{y}_{ij}}$ is the training set label hash code, $sign(W_{j}^{T}{{x}_{i}})$ represents a hash code that generates a lung nodule image, and finally generates a hash database $H=[{{h}_{1}},{{h}_{2}},\cdots ,{{h}_{n}}]$.
3.6. Adam
The Adam algorithm optimizer is a low-moment-based adaptive estimator that updates network parameters. Its advantages are high computational efficiency and low memory requirements. The learning rate of parameters can be dynamically adjusted according to the loss function of the first derivative estimation and the second derivative estimate of each parameter gradient to achieve the optimal solution. Adam is based on the gradient descent method, but the parameters of each iteration of the learning step have a certain range, not because of the large gradient that leads to a large learning step, but because the value of the parameter is relatively stable. It is shown in Equation (16):
In Equation (17),$f({{\theta }_{t}})$ is the objective function, In Equation (20),${{m}_{t}}$ and ${{\hat{m}}_{t}}$ are the deviations and offset correction estimates at the first derivative. In Equation (21),${{v}_{t}}$ and ${{\hat{v}}_{t}}$ are the deviations and offset correction estimates at the second derivative.In Equation (18) and In Equation (19),${{\beta }_{1}}$ and ${{\beta }_{2}}$ are the corresponding decay rates, and add the same weight Attenuation term. The default value of the parameters are ${{\beta }_{1}}$ = 0.9, ${{\beta }_{2}}$= 0.999, $\varepsilon \text{=1}{{\text{0}}^{\text{-8}}}$.
4. Experiments
4.1. Middle Layer Retrieval
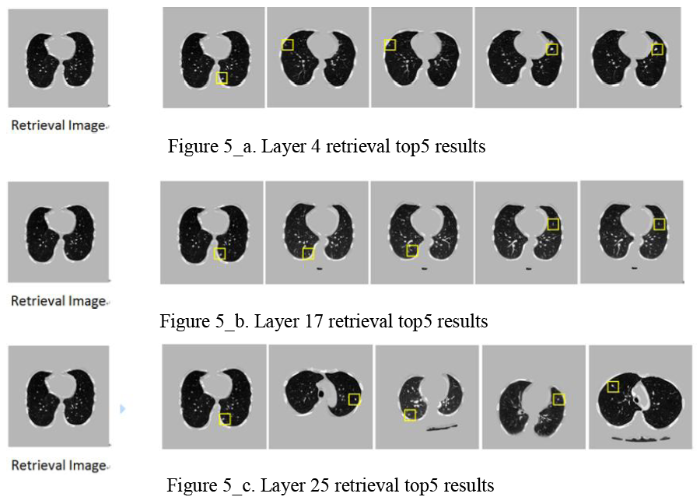
The experiment was performed according to the given label in the center of the lung nodules and 3 slices were used in each case. The lung image was obtained by pre-processing each slice as shown in Figure 1. Through the FCDN network structure, encoding feature extraction and decoding layer up Sampling obtained lung nodule image features. Experiments on all network layers were compared to top5 with the highest retrieval performance.The Figure 5_a, Figure 5_b, and Figure 5_c shows the results of three network middle layers with the smallest Hamming distance:
Figure 5.
Figure 5.
a. Layer 4 retrieval top5results;b. Layer 17 retrieval top5 results;c. Layer 25 retrieval top5 results
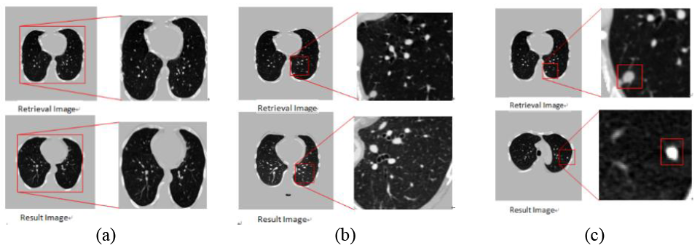
In the experimental results shown above, it is found that the superficial layer of the network extracts more contour information of the whole image, as shown in Figure 6(a) network layer 4 search image information contrast. With the deepening of the network level, extracting more global semantic features of lung nodules images, as shown in Figure 6(b) comparison of the 17th layer search amplification. The final decoding layer of the network is more abundant in the shape and size information of the lung nodules, as shown in Figure 6(c), the network retrieves the lung nodules in the 25th layer. This local feature still does not express the complete semantic information of the image.
Figure 6.
Figure 6.
Network middle layer retrieval results feature comparison
4.2. Joint Distance
The experimental results on the Figure 6prove that the features extracted by different network layers have different meanings. In order to improve the accuracy, the image semantic difference of lung nodules is reduced. The Joint distance was proposed to extract the salient features of lung nodules as the objective function. The global contour of the image features and the global semantic features of the lung nodules were selected as regularization. The equation is
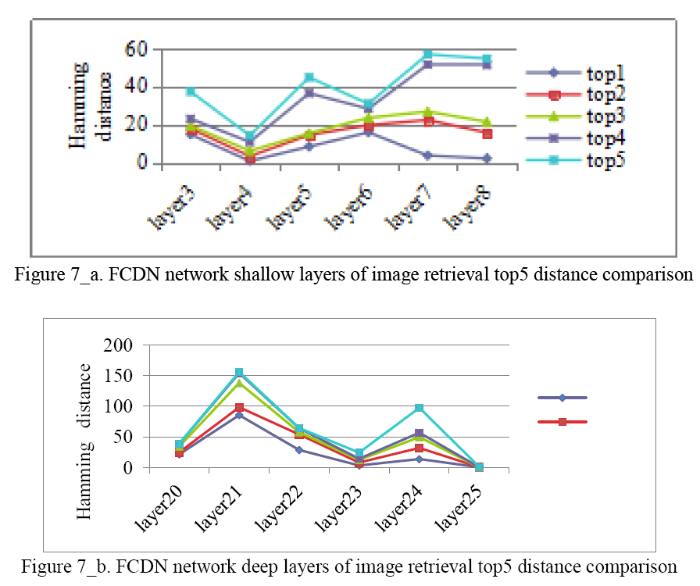
In Equation (22), Discom represents the final Hamming distance andmin(DL) represents the minimum shallow layer Hamming distance, the most image contour information. DM represents the last layer of encoding, which is the most abundant semantic layer of lung nodules. min(DD)represents the minimum deep layer Hamming distance, which is similar to the size of lung nodules. In the experiment, we output encoding and decoding top5 lung nodules image retrieval Hamming distance comparison. This is shown in Figure 7_a and Figure 7_b.
Figure 7.
Figure 7.
a. FCDN network shallow layers of image retrieval top5 distance comparison;b. FCDN network deep layers of image retrieval top5 distance comparison
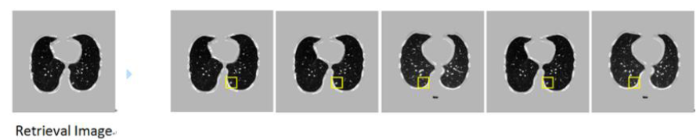
It can be seen from the figure above, FCDN encoding has a shallow selection 4th layer.Decoding deep selection 25th layer and encoding the last layer of 17th layer forms a Joint distance to sort the returnof the lung nodule image with the highest similarity. Joint distance lung nodule image retrieval top5 results are shownin Figure 8.
Figure 8.
Figure 8.
Combine distance retrieval top5 result
The retrieval results onthe Figure 8prove that the proposed Joint distance effectively solves the problem of semantic differences in retrieval image features. The retrieval results are similar to those of retrieval image semantic information and lung nodulesfeatures.
4.3. The Evaluation of the Retrieval Performance
This article uses the most commonly used evaluation standard in the field of image retrieval for final assessment of results: respectivelyprecision, recall, and Mean Average Precision (MAP). The precision refers to the ratio of the number of similar images returned by a query to the total number of returned images. The more correct similar images, the higher the precision of the search. The recall rate indicates the proportion of the number of similar images in the returned result to the number of all similar images in the database.The mathematical expressions are shown Equation (23) and Equation (24).
First, the lung image retrieval hash code in the paper consists of different network layers of Joint distance. Table 1 compares the MAP values of this paper’s method and other hash methods. In Table 1, ABR is an adaptive bit retrieval algorithm proposed in [20]. The average accuracy of our method in 64 bits length hash search is 91.7%, which is better than other methods.
Table 1. Other methods of different bit MAP (%) value
| Method | 24bit 32 bit 48 bit 64 bit |
|---|---|
| ours | 76.1 78.3 86.7 91.7 |
| ABR | 69.6 74.8 78.7 88.2 |
| KSL | 66.2 70.8 74.5 75.9 |
| ITQ | 55.9 66.7 72.5 76.3 |
| SH | 26.4 33.4 36.1 35.8 |
| LSH | 10.3 18.2 29.1 36.5 |
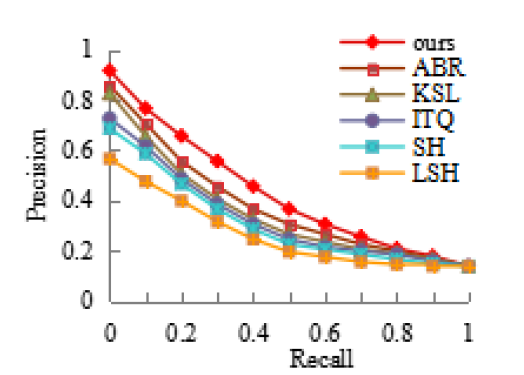
In addition, Figure 9 shows the relationship between the precision and recall of different hash methods under a 64-bit hash code.
Figure 9.
Figure 9.
Comparisons of precision and recall of lung nodule retrieval
5. Conclusion
The retrieval of medical images is of great significance to the assistant diagnosis and treatment of doctors.Early detection of problems with traditional lung cancer methods.This paper proposes a network structure of FCDN, which improves the features extraction that cannot express the image semantic problems. Using the characteristics of different information in different layers of the network to express the images, the minimum Joint distance was used to search the image generated hash codes. The retrieval results made the image semantics and lung nodule features more abundant and improved the retrieval precision.Retrieval of one lung slice requires an average of $4.8\times {{10}^{-5}}$s, which is about 104 times more efficient than direct retrieval of high-dimensional vectors. Another key technology to improve medicalimage retrieval performance is the choice of using a hash function. The direction to explore is how to choose a hashing function with high learning ability.
Acknowledgements
This work is partially supported by Shanxi Nature Foundation (No. 2015011045). The authors also gratefully acknowledge the helpful comments and suggestions of the reviewers, which have improved the presentation.
Reference
“Medical Image Retrieval using Deep Convolutional Neural Network, ”
ImageNet Classification with Deep Convolutional Neural Networks, ”
in
“Very Deep Convolutional Networks for Large-Scale Image Recognition, ”
“Sharing Residual Units Through Collective Tensor Factorization in Deep Neural Networks, ”
in
“Deep Residual Learning for Image Recognition, ”
in
“Accurate Image Super-Resolution using Very Deep Convolutional Networks, ”in
“Distinctive Image Features from Scale-Invariant Keypoints, ”
Densely Connected Convolutional Networks, ”
in
“Learning Multiple Layers of Features from Tiny Images,
”
“ImageNet: A Large-Scale Hierarchical Image Database, ”
in
Histograms of Oriented Gradients for Human Detection, ”
in
“Identity Mappings in Deep Residual Networks, ”
in
“Deep Hash Learning for Efficient Image Retrieval
”in
“Deep Residual Hashing, ”
“Deep Hashing based Fusing Index Method for Large-Scale Image Retrieval
”
“Deep Networks with Stochastic Depth, ”
in
“Cnn Features off-the-Shelf: An Astounding Baseline for Recognition
” in
“Region-based Convolutional Networks for Accurate Object Detection and Semantic Segmentation, ”
“Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift
, ”
“Hashing Retrieval for CT Images of Pulmonary Nodules based on Medical Signs and Convolutional Neural Networks, ”